In biology, as in real estate, location matters. Working copies of active genes — called messenger RNAs or mRNAs — are positioned strategically throughout living tissues, and their location often helps regulate how cells and tissues grow and develop. But to analyze many mRNAs simultaneously, scientists have had to grind cells to a pulp, which left them no good way to pinpoint where those mRNAs sat within the cell.
Now a team at the Wyss Institute of Biologically Inspired Engineering at Harvard University and Harvard Medical School, in collaboration with the Allen Institute for Brain Science, has developed a new method that allows scientists to pinpoint thousands of mRNAs and other types of RNAs at once in intact cells — all while determining the sequence of letters, or bases, that identify them and reveal what they do.
The method, called fluorescent in situ RNA sequencing (FISSEQ), could lead to earlier cancer diagnosis by revealing molecular changes that drive cancer in seemingly healthy tissue. It could track cancer mutations and how they respond to modern targeted therapies, and uncover targets for safer and more effective ones.
The method could also help biologists understand how tissues change subtly during embryonic development — and even help map the maze of neurons that wire the human brain. The researchers reported the method in today’s online edition of Science.
“By looking comprehensively at gene expression within cells, we can now spot numerous important differences in complex tissues like the brain that are invisible today,” said George Church, Ph.D., a Core Faculty member at the Wyss Institute and Professor of Genetics at Harvard Medical School. “This will help us understand like never before how tissues develop and function in health and disease.”
Locking RNAs in Place
Healthy human cells typically turn on nearly half of their 20,000 genes at any given time, and they choose those genes carefully to produce the desired cellular responses. Moreover, cells can dial gene expression up or down, adjusting to produce anywhere from a few working copies of a gene to several thousand.
But simultaneously pinpointing the cellular location of all those mRNAs is a tall order.
Church and Je Hyuk Lee, Ph.D., a Research Fellow at the Wyss Institute and Harvard Medical School, were up for the challenge. Moreover, they wanted to simultaneously determine the sequence of those RNAs, which identifies them and often reveals their function.
Lee and his colleagues first treated the tissue chemically to fix the cell’s thousands of RNAs in place. Then they used enzymes to copy those RNAs into DNA replicas, and copy those replicas many times to create a tiny ball of replica DNA fixed to the same spot.
They managed to fix and replicate thousands of the cell’s RNAs at once — but then became a victim of their own success. The RNAs were so tightly packed inside the cell that even a tricked-out microscope and camera could not distinguish the flashing lights of one individual ball of replica DNA from those of its neighbors.
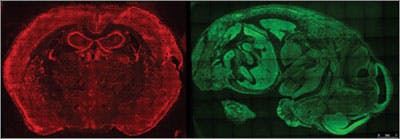
A new way to image RNA
To solve that problem, the researchers pioneered an unconventional method to visualize tiny objects inside cells. It works like an urban postal system. If a postmaster tried to identify each home in her city by color, she would quickly run out of colors as new homes were built, resulting in undelivered mail. Instead, postmasters keep track of each home by assigning it a unique address.
The researchers realized they could assign each RNA in the cell a unique address: the sequence of “letters,” or bases, in the RNA molecule itself. And they figured they could read the address using methods akin to next-generation DNA sequencing, a set of high-speed genome sequencing methods Church helped develop in the early 2000s.
In next-gen sequencing, scientists grind up tissue, extract its DNA, break the DNA into pieces, then dilute those pieces enough so that each piece of DNA sticks to a separate spot on a glass slide. They use enzymes and four different fluorescent dyes — one each for each of the four “letters,” or bases — to make the DNA flash a sequence of colors that reveals its sequence.
By analogy, the scientists sought to fix RNA in place in the cell, make a tiny ball with many matching DNA replicas of each RNA, then adapt next-gen DNA sequencing so it worked in fixed cells. The four flashing colors would reveal the base sequence of each replica DNA, which would tell them the base sequence of the matching RNA from which it was derived. And those sequences would in theory provide an unlimited number of unique addresses — one for each of the original RNAs.
The scientists struggled at first to visualize the flashing lights of individual balls of replica DNA from a distance where the whole landscape of the tissue remained in view. They succeeded by selectively turning on just a fraction of those flashing dots at any given time, so they could distinguish single balls of replica DNA flashing across the cellular landscape.
The strategy would only work, however, if they could actually read enough of the base sequence to provide a unique address. At first they could not determine more than six bases in the replica DNA, which did not provide enough unique addresses to identify individual genes in the human genome. That’s when Evan Daugharthy, a graduate student at Harvard Medical School, stepped in.
FISSEQ at Work
Daugharthy first devised an algorithm to locate the sequence of the replica DNA with the known sequence of genes in the human genome. Flashing lights that did not correspond to a real gene were erased from the image.
Then Daugharthy hacked a commercial DNA sequencing kit, which enabled the team to sequence 30 bases, more than enough to provide each replica DNA with a unique address. In this way the team could create a composite image representing the sequence, and location, of RNA corresponding to every gene in the human genome.
Lee, Daugharthy and their colleagues then tested the method to detect the genes skin cells turn on as they multiply and migrate to heal a simulated wound in a petri dish. Cells growing into the wound had 12 genes that were activated much more or much less than nearby cells sitting idly on the sidelines. Similar experiments could identify new markers of diseased tissue or new targets for targeted molecular therapies.
“What George’s team has accomplished is a technological tour de force,” said Wyss Institute Founding Director Don Ingber, M.D., Ph.D. “By spotting incredibly subtle but incredibly important changes in gene expression and precisely defining their position inside the cell, they have helped open the door to a new age of cellular diagnostics.”
The work was funded by the National Institutes of Health, the Allen Institute for Brain Science and the Wyss Institute.