New technology could launch biomedical imaging to next level
BOSTON — Much like the checkout clerk uses a machine that scans the barcodes on packages to identify what customers bought at the store, scientists use powerful microscopes and their own kinds of barcodes to help them identify various parts of a cell, or types of molecules at a disease site. But their barcodes only come in a handful of "styles," limiting the number of objects scientists can study in a cell sample at any one time.
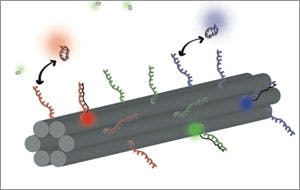
Researchers at the Wyss Institute for Biologically Inspired Engineering at Harvard University have created a new kind of barcode that could come in an almost limitless array of styles — with the potential to enable scientists to gather vastly more vital information, at one given time, than ever before. The method harnesses the natural ability of DNA to self-assemble, as reported today in the online issue of Nature Chemistry.
"We hope this new method will provide much-needed molecular tools for using fluorescence microscopy to study complex biological problems," says Peng Yin, Wyss core faculty member and study co-author who has been instrumental in the DNA origami technology at the heart of the new method.
Fluorescence microscopy has been a tour de force in biomedical imaging for the last several decades. In short, scientists couple fluorescent elements -— the barcodes — to molecules they know will attach to the part of the cells they wanted to investigate. Illuminating the sample triggers each kind of barcode to fluoresce at a particular wavelength of light, such as red, blue, or green — indicating where the molecules of interest are.
However, the method is limited by the number of colors available — three or four — and sometimes the colors get blurry. That’s where the magic of the DNA barcode comes in: colored-dots can be arranged into geometric patterns or fluorescent linear barcodes, and the combinations are almost limitless — substantially increasing the number of distinct molecules or cells scientists can observe in a sample, and the colors are easy to distinguish.
Here’s how it works: DNA origami follows the basic principles of the double helix in which the molecular bases A (adenosine) only bind to T (thymine), and C (cytosine) bases only bind to G (guanine). With those "givens" in place, a long strand of DNA is programmed to self-assemble by folding in on itself with the help of shorter strands to create predetermined forms—much like a single sheet of paper is folded to create a variety of designs in the traditional Japanese art.

To these more structurally complex DNA nano-structures, researchers can then attach fluorescent molecules to the desired spots, and use origami technology to generate a large pool of barcodes out of only a few fluorescent molecules. That could add a lot to the cellular imaging "toolbox" because it enables scientists to potentially light up more cellular structures than ever possible before.
"The intrinsic rigidity of the engineered DNA nanostructures is this method’s greatest advantage; it holds the fluorescent pattern in place without the use of external forces. It also holds great promise for using the method to study cells in their native environments," Yin says. As proof of concept, the team demonstrated that one of their new barcodes successfully attached to the surface of a yeast cell.
More research beckons, particularly to determine what happens when each of the fluorescent barcodes are mixed together in a cell sample, which is routine in real-life biological and medical imaging systems—but there’s plenty of good news as a starting point. It’s low-cost, easy to do, and more robust compared to current methods, says Yin.
"We’re moving fast in our ability to manipulate DNA molecules using origami technology," says Wyss Institute Founding Director Don Ingber, M.D., Ph.D., "and the landscape of its potential is tremendous — from helping us to develop targeted drug-delivery mechanisms to improving the scope of cellular and molecular activities we are able to observe at a disease site using the latest medical imaging techniques."
The research team was led by three Wyss Founding Core Faculty members: Peng Yin, Ph.D., William Shih, Ph.D., and George Church, Ph.D. Yin is also an Assistant Professor of Systems Biology at Harvard Medical School (HMS). Church is also Professor of Genetics at HMS and Professor of Health Sciences and Technology at Harvard and the Massachusetts Institute of Technology. Shih is an Associate Professor in the Department of Biological Chemistry and Molecular Pharmacology at HMS and the Department of Cancer Biology at the Dana-Farber Cancer Institute. Other research contributors included Chenxiang Lin, Ph.D., now Assistant Professor of Cell Biology at Yale School of Medicine; Wyss Institute Postdoctoral Scholar Ralf Jungmann, Ph.D.; Wyss Staff Scientist Chao Li, Ph.D.; Wyss Senior Staff Scientist Daniel Levner, Ph.D.; and Andrew Leifer, Ph.D., formerly at Harvard University, who is now a Lewis-Sigler Fellow at Princeton University.
The work was funded by the National Institutes of Health, the National Science Foundation, the Office of Naval Research, and the Wyss Institute.